| Nucleic acids are polymers of nucleotides. Each nucleotide includes a base, either a purine (adenine, guanine) or apyrimidine (cytosine, uracil, thymine). See diagram at right . Some nucleic acids contain modified bases. |  |
| In a nucleotide, e.g., adenosine monophosphate (AMP), the base (e.g., adenine) is bonded to a ribose sugar, which in turn has a phosphate in ester linkage to the 5' hydroxyl, as shown at right. |  |
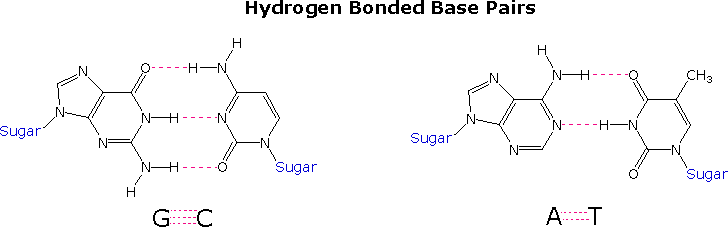
Hydrogen bonds link two complementary nucleotide bases on separate nucleic acid strands, or on complementary portions of the same strand. The conventional base pairs are between A & U (or T) and between C & G, as shown in Fig. 5-12 p.88 and below.
 |
In the diagram at left, H-bonds are shown in red. Relative bond lengths are inexact.
The image at right is based on X-ray crystallography of tRNAGln (PDB file 1GTR). H atoms are not shown.
|  |
Secondary structure: Base pairing over extended stretches of complementary base sequences in two nucleic acid strands stabilizes secondary structure, such as the double helix of DNA (p. 87, 1108). Stacking interactions between adjacent hydrophobic bases in the double helix, contribute to stabilization of such secondary structures. Each base interacts with its neighbors above and below, in the ladder-like arrangement of base pairs in the double helix, e.g., of DNA.
DNA
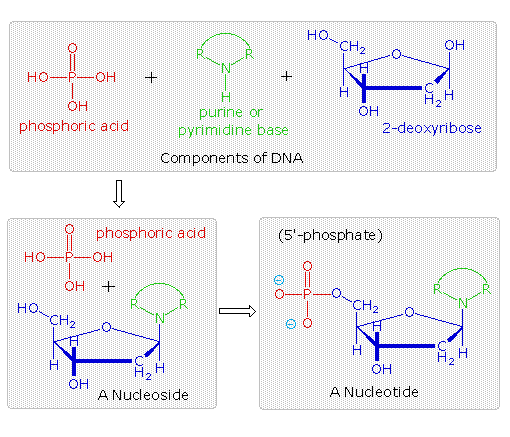
DNA is a polymer, consisting of monomers call deoxynucleotides. The monomer contains a simple sugar (deoxyribose, shown in black below), a phosphate group (in red), and a cyclic organic R group (in blue) that is analogous to the side chain of an amino acid.
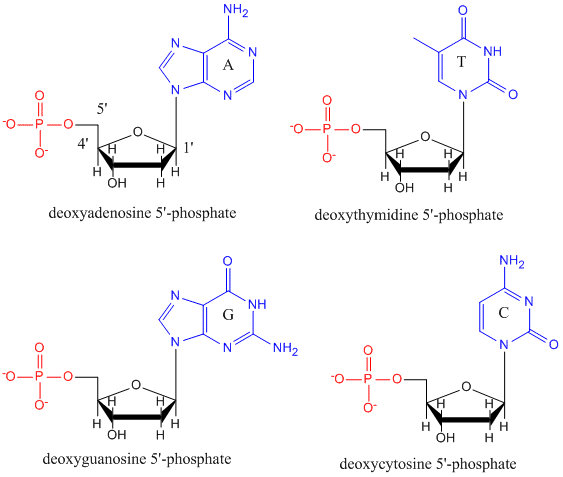
Only four bases are used in DNA (in contrast to the 20 different side chains in proteins) which we will abbreviate, for simplicity, as A, G, C and T. They are bases since they contain amine groups that can accept protons.
The polymer consists of a sugar - phosphate - sugar - phosphate backbone, with one base attached to each sugar molecule.
As with proteins, the DNA backbone is polar but also charged. It is a polyanion.
The bases, analogous to the side chains of amino acids, are predominately polar.
Given the charged nature of the backbone, you might expect that DNA does not fold to a compact globular (spherical) shape, even if positively charged cations like Mg bind to and stabilize the charge on the polymer.
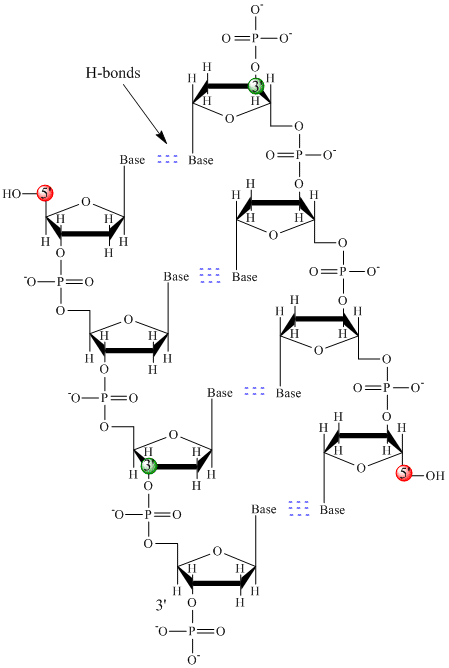
Instead, DNA exists usually as a double-stranded (ds) structure with the sugar-phosphate backbones of the two different strands running in opposite directions (5'-3' and the other 3'-5').
The strands are held together by hydrogen bonds between bases on complementary strands.
Hence like proteins, DNA has secondary structure but in this case, the hydrogen bonds are not within the backbone but between the "side chain" bases on opposing strands.
It is actually a misnomer to call dsDNA a molecule, since it really consists of two different, complementary strands held together by hydrogen bonds. A structure of ds-DNA showing the opposite polarity of the strands is shown below.
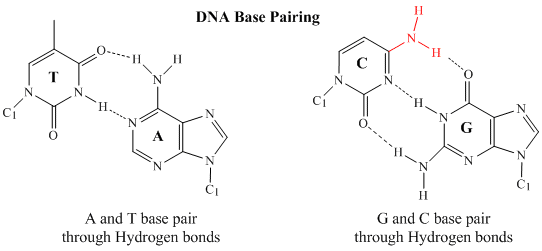
 In double stranded DNA, the guanine (G) base on one strand can form three H-bonds with a cytosine (C) base on another strand (this is called a GC base pair). The thymine (T) base on one strand can form two H-bonds with an adenine (A) base on the other strand (this is called an AT base pair). Double-stranded DNA has a regular geometric structure with a fixed distance between the two backbones. This requires the bases pairs to consists of one base with a two-ring (bicyclic) structure (these bases are called purines) and one with a single ring structure (these bases are called pyrimidines). Hence a G and A or a T and C are not possible base pair partners.
In double stranded DNA, the guanine (G) base on one strand can form three H-bonds with a cytosine (C) base on another strand (this is called a GC base pair). The thymine (T) base on one strand can form two H-bonds with an adenine (A) base on the other strand (this is called an AT base pair). Double-stranded DNA has a regular geometric structure with a fixed distance between the two backbones. This requires the bases pairs to consists of one base with a two-ring (bicyclic) structure (these bases are called purines) and one with a single ring structure (these bases are called pyrimidines). Hence a G and A or a T and C are not possible base pair partners.


The Chemical Nature of DNA
The polymeric structure of DNA may be described in terms of monomeric units of increasing complexity.
In the top shaded box of the following illustration, the three relatively simple components mentioned earlier are shown.
Below that on the left , formulas for phosphoric acid and a nucleoside are drawn. Condensation polymerization of these leads to the DNA formulation outlined above.
Finally, a 5'- monophosphate ester, called a nucleotide may be drawn as a single monomer unit, shown in the shaded box to the right.
Since a monophosphate ester of this kind is a strong acid (pKa of 1.0), it will be fully ionized at the usual physiological pH (ca.7.4).
Names for these DNA components are given in the table to the right of the diagram. Isomeric 3'-monophospate nucleotides are also known, and both isomers are found in cells.
They may be obtained by selective hydrolysis of DNA through the action of nuclease enzymes.
Anhydride-like di- and tri-phosphate nucleotides have been identified as important energy carriers in biochemical reactions, the most common being ATP (adenosine 5'-triphosphate).
Several important characteristics of this formula should be noted.
• First, the remaining P-OH function is quite acidic and is completely ionized in biological systems.
• Second, the polymer chain is structurally directed. One end (5') is different from the other (3').
• Third, although this appears to be a relatively simple polymer, the possible permutations of the four nucleosides in the chain become very large as the chain lengthens.
• Fourth, the DNA polymer is much larger than originally believed. Molecular weights for the DNA from multicellular organisms are commonly 109 or greater.
• Second, the polymer chain is structurally directed. One end (5') is different from the other (3').
• Third, although this appears to be a relatively simple polymer, the possible permutations of the four nucleosides in the chain become very large as the chain lengthens.
• Fourth, the DNA polymer is much larger than originally believed. Molecular weights for the DNA from multicellular organisms are commonly 109 or greater.
The Secondary Structure of DNA
Base Pairing
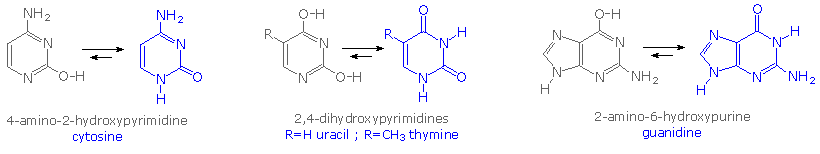
Careful examination of the purine and pyrimidine base components of the nucleotides reveals that three of them could exist as hydroxy pyrimidine or purine tautomers, having an aromatic heterocyclic ring. Despite the added stabilization of an aromatic ring, these compounds prefer to adopt amide-like structures. These options are shown in the following diagram, with the more stable tautomer drawn in blue.
Careful examination of the purine and pyrimidine base components of the nucleotides reveals that three of them could exist as hydroxy pyrimidine or purine tautomers, having an aromatic heterocyclic ring. Despite the added stabilization of an aromatic ring, these compounds prefer to adopt amide-like structures. These options are shown in the following diagram, with the more stable tautomer drawn in blue.
The G#C association involves three hydrogen bonds (colored pink), and is therefore stronger than the two-hydrogen bond association of A#T.
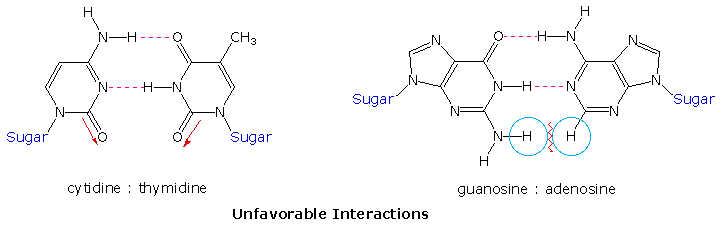
A simple mnemonic device for remembering which bases are paired comes from the line construction of the capital letters used to identify the bases. A and T are made up of intersecting straight lines. In contrast, C and G are largely composed of curved lines. The RNA base uracil corresponds to thymine, since U follows T in the alphabet.
The Double Helix
After many trials and modifications, Watson and Crick conceived an ingenious double helix model for the secondary structure of DNA.
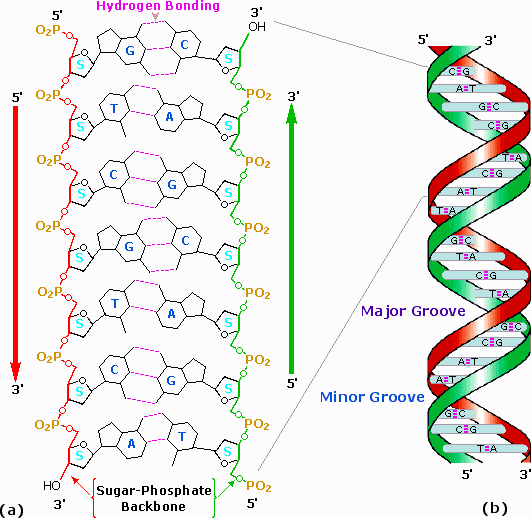
Two strands of DNA were aligned anti-parallel to each other, i.e. with opposite 3' and 5' ends , as shown in part a of the following diagram.
Complementary primary nucleotide structures for each strand allowed intra-strand hydrogen bonding between each pair of bases.
These complementary strands are colored red and green in the diagram.
Coiling these coupled strands then leads to a double helix structure, shown as cross-linked ribbons in part b of the diagram.
The double helix is further stabilized by hydrophobic attractions and pi-stacking of the bases.
A space-filling molecular model of a short segment is displayed in part c on the right.
The helix shown here has ten base pairs per turn, and rises 3.4 Å in each turn. This right-handed helix is the favored conformation in aqueous systems, and has been termed the B-helix.
As the DNA strands wind around each other, they leave gaps between each set of phosphate backbones.
Two alternating grooves result, a wide and deep major groove (ca. 22Å wide), and a shallow and narrow minor groove(ca. 12Å wide).
Other molecules, including polypeptides, may insert into these grooves, and in so doing perturb the chemistry of DNA. Other helical structures of DNA have also been observed, and are designated by letters (e.g. A and Z).
 | Space-Filling Molecular Model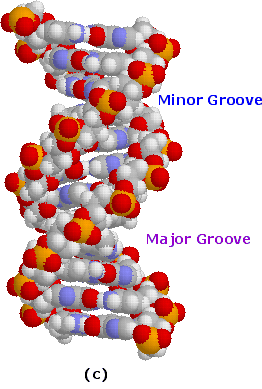 |

No comments:
Post a Comment